The peroxisomal matrix protein import system shares mechanistic similarities with the endoplasmic reticulum/proteasome degradation process (ERAD), indicating a common evolutionary history.
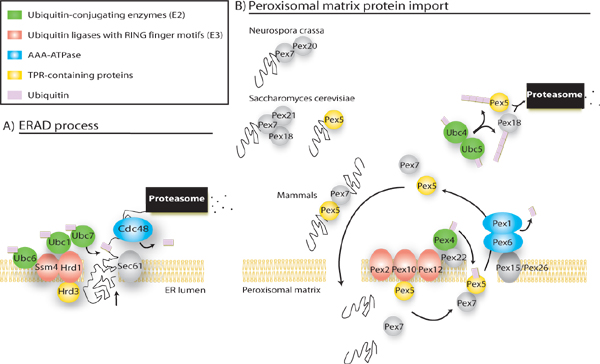
Figure: ER-peroxisome connection a) Endoplasmic reticulum associated degradation process (ERAD), adapted from (McCracken and Brodsky 2003). b) Essential components of the import machinery in peroxisomes. Based on orthology/ phylogenetic criteria, we propose that peroxisome and ER would use a similar mechanism to recycle the peroxisomal receptors and export proteins to the cytosol, respectively. First, the proteins directed to the peroxisome would be binding to the peroxisomal Pex5 or Pex7 receptors (Pex7 binds to Pex20 in N. crassa, to Pex18 and Pex21 in S. cerevisiae, and to the long form of Pex5 in mammals). Once bound to their cargoes, Pex5, and Pex7 would approach to the peroxisomal membrane and get translocated into the peroxisome. In the matrix, receptors will be uncoupled and exported to the cytosol for recycling. The attachment of ubiquitin to Pex5 emerging from the export channel, most likely by Pex4, would drive the receptor dislocation. Pex1 and Pex6 would then recognize this single ubiquitin as signal, then unfold the complex and mediate the export to the cytosol using ATP hydrolysis. Other cytosolic E2, such as Ubc4 and Ubc5 would polyubiquitinate Pex5 and Pex18 and address them to the proteasome for degradation.
A recent work (Gabaldón et al, 2006) show that most peroxisomal proteins (39–58%) are of eukaryotic origin, comprising all proteins involved in organelle biogenesis or maintenance. A significant fraction (13–18%), consisting mainly of enzymes, has an alpha-proteobacterial origin and appears to be the result of the recruitment of proteins originally targeted to mitochondria.
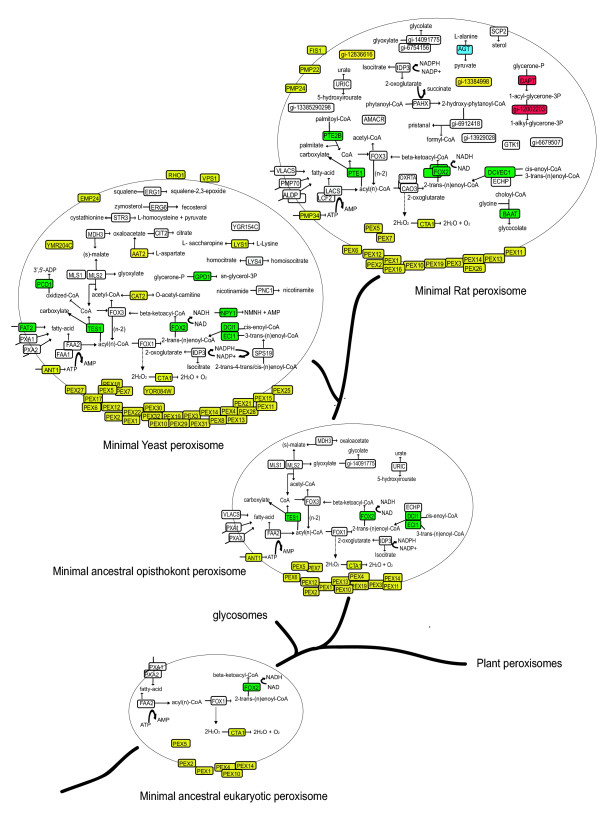
Figure: Evolution of the peroxisomal proteome Evolution of the peroxisomal proteome. Biochemical pathways reconstructed according to KEGG and annotations of peroxisomal proteins. For details on the reconstruction of ancestral states see supplemental material. Color code: yellow, eukaryotic origin; green, alpha-proteobacterial origin; red, actinomycetales origin; blue, cyanobacterial origin; white, origin unresolved. Note that the ancestral eukaryotic peroxisomal proteome reconstruction depends on the topology of the eukaryotic tree. If an alternative topology is considered, placing kinetoplastida and viridiplantae together [49], and the plant peroxisomal proteome is taken from the Araperox database [31], then the reconstructed ancestral eukaryotic peroxisomal proteome would be much larger, including all proteins present in the opisthokont proteome except for ANT1, IDP3, FOX3, PEX13 and PEX19.