Target Signal Predictor
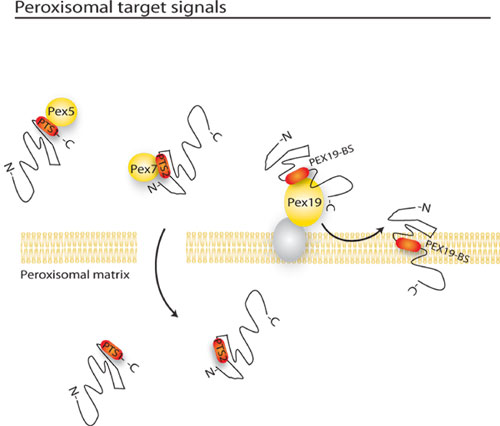
Three targeting sequences for peroxisomal proteins have been identified. Two of them, PTS1 and PTS2, identify peroxisomal matrix proteins; PEX19BS identifies peroxisomal membrane proteins instead. Depending on the type of PTS signal the proteins might carry, they are translocated to the peroxisome via Pex5 (PTS1 receptor), Pex7 (PTS2 receptor) or Pex19 (Membrane protein receptor).
PTS1
PTS1 is the most abundant target signal, and is located in the C-terminal. The PTS1 is highly conserved in evolution, with the following tripeptide consensus S/A-K/R-L/M. Moreover, beyond this tripeptide motif, the PTS1 motif has been recently enlarged up to 12 C-terminal residues
(Neuberger et al, 2003) , comprising:
Our PTS1 BLOCK relies on an alignment of the 12 C-terminal residues of 99 bona fide PTS1 proteins from Homo sapiens, Mus musculus, Saccharomyces cerevisiae and Arabidopsis thaliana. Because the searcher does not discriminate the possible location of the motif along the query sequence (whether C- or N-terminal), identification of the truly C-terminal PTS1 motif must eventually be done manually.
Validation tests:
PTS2
Only three different proteins in Metazoan and fungi carrying this target signal have been identified:
In the rat ACAA1 protein, the PTS2 localises at the 16 N-terminal residues. The arginine at position 4 and the leucines at position 5 and 12 are critical for efficient targeting (Glover et al, 1994). The consensus sequence corresponds to R-L/I-X(5)-HL. Our PTS2 BLOCK relies on the alignment of the ungaped motifs comprising these critical residues of ACAA1, PHYH and AGPS (Homo sapiens, Mus musculus, Ratus norvegicus, Saccharomyces cerevisiae, Kluyveromyces lactis, Arabidopsis thaliana)
Validation tests:
Pex19BS
Membrane proteins are not targeted via PTS1 or PTS2, but via Pex19. Pex19 is a membrane protein that interacts with all Peroxisomal Membrane Proteins (PMPs) for their import. We used the experimental characterisation of the PMPs motif interaction either in
Saccharomyces cerevisiae
(Rottensteiner et al, 2004) or Homo sapiens
(Halbach et al, 2005) in order to make the Pex19BS BLOCK.
MEME
We apply the MEME
system on a selection of manually curated proteins that carry a PTS, for a refinement and identification of key residues. These MEME motifs were submitted to BLOCKS multiple alignment processor.
BLOCKS
Blocks consist of multiply aligned sequence segments without gaps that represent the highest conserved regions. Searches of the PTS Blocks are carried out using "Do-It-YourSelf Block Search" at the
BLOCKS server
. Results are returned with measures of probability.
Links to other PTS predictors